Motivation
Accurate inference of genealogical relationships between pairs of individuals is paramount in association studies, forensics and evolutionary analyses of wildlife populations. Current methods for relationship inference consider only a small set of close relationships and have limited to no power to distinguish between relationships with the same number of meioses separating the individuals under consideration (e.g. aunt-niece vs niece-aunt or first cousins vs great aunt-niece). In association studies, these limitations can lead to unnecessary exclusions of putative relatives.
CARROT
CARROT (ClAssification of Relationships with ROTations) is our novel framework for relationship inference that leverages linkage information to differentiate between rotated relationships, that is, between relationships with the same number of common ancestors and the same number of meioses separating the individuals under consideration.
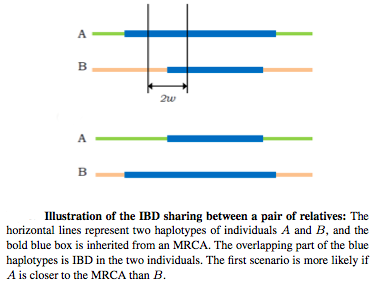
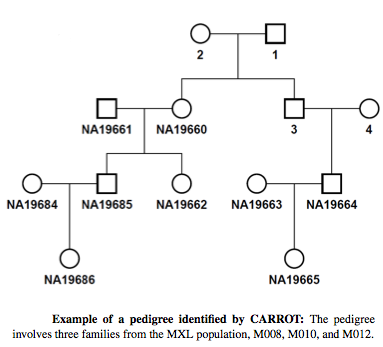
We demonstrated that CARROT clearly outperforms existing methods on simulated data. We also applied CARROT on four populations from Phase III of the HapMap Project and detected previously unreported pairs of third and fourth degree relatives.
Download and Contact
The sources for CARROT are publicly and freely available
here.
For further information, please contact Sofia Kyriazopoulou-Panagiotopoulou at sofiakp AT stanford DOT edu, or
Serafim Batzoglou's lab.



